|
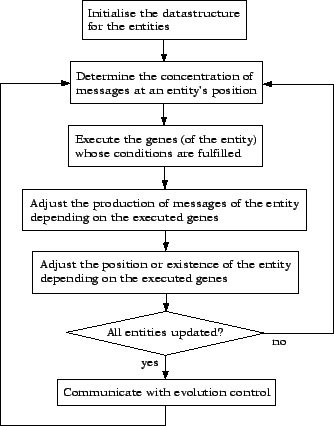
A simple multicellular program works as shown in figure 2.1. First, the cells (here named entities because even though they communicate like cells they normally represent something else like for example robots or sensory units or simply program modules) have to be instantiated from the genome and stored into a datastructure that can easily be run through for updating the cells. In the current implementation (which is written in C++ and two-dimensional) this datastructure is a map of maps which contain the cells. As the cells are instantiations of the class Genome, this datastructure is declared as follows:
The key values of the maps contain the position of the cell. This is not necessarily so. If we would have entities that move (e.g. robots) it would probably be a better idea to not care about the keys and only change the position stored in the entities themselves.
After this initialization, the main function runs through the cells datastructure and calls the update() function of each cell. This function first computes the total concentration of messages20 at the cell's position. For that purpose it runs through all cells and adds the influence of their message productions on the local concentration. This influence is determined on the basis of the distance between the message producer and the updated cell and can include arbitrary variations by a modeled environment in the virtual space between the two cells. The calculation of the influence of a produced message on the local concentration is taken care of by a function called howMuch() that has access to the simulated environment.
In the next step, the update() function calls all the genes of the cell. The genes themselves test all their conditions for activation before executing their evolved program code and producing a message. This ensures that only genes whose activation conditions are met get executed.
After that, the update() function first updates the message production of the current cell depending on which of its genes got executed (and therefore could produce a message). Then, it adjusts the position (in the case of moving entities) or the existence of the entity (in the case of cell death) or even of other entities (in the case of cell division). This could also be done by the genes directly, but as these are important actions for the functioning of the individual, they are presented as separate steps in figure 2.1.
The communication with the part of the system that takes care of the evolution of good individuals21 can be done after each single update or even directly in the genes, but here it is assumed that most of this communication happens after all the cells have been updated22.
The OOOP individual (i.e. the multicellular program) consists of the following parts:
The class Genome consists of the following parts:
A gene consists of the following parts: