|
The Object-Oriented Ontogenetic Programming System is an ensemble of programs which work together for breeding multicellular programs. The underlying techniques are object-oriented ontogenetic programming for the multicellular programs and genetic programming for the breeding. For coping with the problems of OOOP and GP that are described in sections 2.1.4 and 1.5.2, OOOPS introduces some extensions to GP. First, we will look at the architecture of the system and its functioning. In section 3.2.2, we will then discuss the extensions of OOOPS to the basic GP approach shown in section 3.1.1.
It is impossible to describe OOOPS in all detail here, because it is such a big system that its complete description would fill a whole book of its own. This section tries to give at least a good overview of the main features and workings of the Object-Oriented Ontogenetic Programming System. Because of limited space, there are also several unexplained technical terms which are common knowledge for programmers.
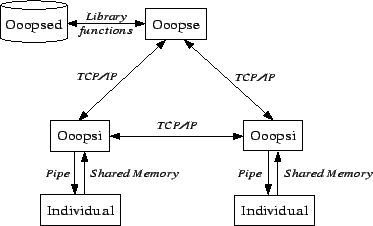
As was already mentioned in section 2.1.4, the Object-Oriented Ontogenetic Programming System is designed such that it can run in parallel, distributed on several computers. For allowing this, OOOPS is organized as a client-server architecture. There are three different programs that have been created to build OOOPS:
Figure 3.3 shows these different parts of OOOPS and their interactions. All parts are described in more detail in the following.
Ooopsed is a relational database which stores all important information about the evolutionary run. As database management system, PostgreSQL is used because it is free and has a lot of practical features. Plus, there are many developers and a good documentation. The database has the following structure:
The parent table is needed for storing information about offspring. As OOOPS works asynchronously9 and therefore only can vary or select one individual at a time, offspring cannot be inserted into the population at once when an individual is selected for reproduction. A reproducing individual does not die. But a new individual can only replace a died individual and take its address. So the children have to wait in the database (in the parent table) until another individual dies and they can take its place. The indid is the ID of the parent individual, numdescs contains the number of children it is allowed to produce, code is the sourcecode of the parent individual and results contains the evaluation results which made Ooopse select this individual for reproduction. When an individual dies, a random child is chosen from the parent table to take its place.
Ooopse is the central server program which controls the evolutionary run by initializing new individuals, taking care of the Ooopsed database, deciding on selection and variation and organizing the reproduction. Ooopse is divided into several modules that take on different parts of the work. As the whole system is implemented in object-oriented manner with the C++ programming language, these modules are realized as different objects. Figure 3.4 gives an overview of the architecture of the Ooopse program.
The server() function is the central module which is constantly called by the main() function. It answers requests coming from Ooopsis by forking the Ooopse program10 and calling functions of the other modules. The server() gets requests from an Ooopsi and answers them through the Socket. More precisely, there are two sockets, a ServerSocket which listens for incoming requests and a ClientSocket with which the Ooopse could request the source code of a specific individual from its hosting Ooopsi. This is not needed for the functioning of the system, but it allows the user at the central computer to get the sourcecode of any individual that is registered in the database.
Ecomm contains all the other functions necessary for communicating with the Ooopsis, for example functions for converting information which is to be sent into a serial format and back to the original datastructures.
UI is meant to be the object controlling the user interface. As there is nearly no user interaction until now, its only function is to save statistical data to the harddisk and draw graphs by sending the data to gnuplot11 through a pipe.
Dcomm is the most important of the Ooopse modules. It not only contains functions for saving the results of individual evaluations to the database, but it also includes all functions that decide on what to do with each single individual. So this module also decides on selection and variation. It makes sense not to construct separate objects for these functions because they must operate directly on the database. All decisions are based on the information contained in the database and the functions have to return information from the database as for example the address of the donor individual for gene conjugation. But except deleting an individual from the database when it dies, Dcomm only decides but does not execute its decisions itself. The decisions are sent to the Ooopsi for execution. Later in this section, we will see more exactly how the different parts of OOOPS work together by looking at the control flow of a typical evolutionary run.
The Ooopsi is a client program that contains the sourcecode of some individuals of the population and takes care of everything concerning these individuals. It compiles, runs and evaluates one individual after the other, sends the results to the Ooopse and then executes the commands (e.g. concerning variation) received as an answer. Thereafter, it compiles, runs and evaluates again. The main parts of the Ooopsi program are shown in figure 3.5.
In the Ooopsi, the main() function not only calls the server() function, but also manages all the other activities. The server() function here is only used for answering requests for individuals or genes that are managed by the Ooopsi. These requests can either come from another Ooopsi (which is the standard for gene conjugation) or from the Ooopse. As in the Ooopse, the server() function uses the Socket object for communication via TCP/IP. It also has access to the Code module where the sourcecodes are stored.
The Code module not only stores the sourcecode of the individuals but it also compiles and varies them based on the instructions received from the Ooopse. It can probably be seen as the most important part of the Ooopsi. It takes care of everything related to the program code of the individuals. Especially complex and important are the different possibilities for variation. They are described separately later in this section.
The Icomm module handles the communication with the individual. So in contrast to the Code object which manages the individual code including the compilation, the Icomm object is in charge of the individual executable. It starts and stops the runs of the individual, sends other instructions (which are mostly problem specific) through a pipe and reads information from the individual through shared memory. This information can for example be the current state of the individual for visualization of its dynamics in a user interface, it can be reactions of the individual which are fed back into the external problem environment managed by the Problem module, or it can be outputs of the individual that are used for evaluating its fitness.
The Problem module is only used for some types of problems. If OOOPS is used for the evolution of distributed intelligence where the problem solution is only the genome of the units and the problem environment is modelled in the individual, the Problem module is not used as there is no external problem environment. But if OOOP is used as a modularization technique and the problem solution is the whole individual, the problem environment is located outside of the individual. In this case, the problem environment will usually be read by the main() function (using the Problem object) which will then feed the environmental inputs into the individual through the Icomm object. These environmental inputs can for example be fed into the individual by varying the message production of special external message sources that can be arbitrarily located in the cell space12. Like this, usual GP training- and testcases can be fed into the individual13.
The Ecomm module does in the case of the Ooopsi not only serve as function package for conversion of message formats used for the communication between the Ooopse and the Ooopsis. Except for the server() function which directly uses the Socket object14, the Ooopsi does not directly access the Socket but communicates through the Ecomm module. So this module handles the whole communication with the other Ooopsis and the Ooopse except answering gene or individual requests.
The individual has already been described in section 2.2.
Selection in OOOPS is carried out by the Ooopse15. First, the individual is ranked by its fitness16. Depending on this rank, the individual can be selected to die, in which case it is neither a candidate for reproduction nor (of course) for variation. There is a parameter mortalPercentage that defines which percentage of the population has a death probability greater than zero. The individuals that appertain to the mortal part of the population are those that have the lower fitness ranks. The individual with the lowest fitness rank has a death probability of almost 117 and this probability decreases linearly with increasing fitness rank until it reaches 0 at the first individual that belongs to the immortal part of the population.
If the individual does not die, the Dcomm.select() function decides whether the individual is to be varied itself or if it is part of the elite which continues unvaried into the next generation. There is a parameter elitistNum which defines how many individuals belong to the elite. An individual remains unvaried in the population if it ranks among the elitistNum fittest individuals.
At last, the Dcomm.select() function decides whether the individual is allowed to produce a child. The individual's reproduction probability is computed analogous to its death probability but with the difference that the probability is 1 for the fittest individual and it decreases linearly with decreasing fitness rank until the first infertile individual is reached. As every child can only replace a dead individual, for not producing too many children the average reproduction probability should be about the same as the average death probability. Therefore, numFertiles is set equal to numMortals. As described earlier in this section, children are not produced directly but noted in the database until they can replace a died individual. Because the parent will probably not stay unvaried until the child is used, the code of the parent is transferred and saved to the database when it is selected for reproduction. This not only ensures that the child will not be produced from a parent that has already lost its good qualities (or has even died) but it also means that there are some copies of the fittest individuals in the database most of the time. The user can take a look at them at every stage of the evolutionary run without requesting separate transmissions18.
Variation in OOOPS is handled both by the Ooopse and by the Ooopsis. The Ooopse function Dcomm.vary() decides on which type of variation is to be executed on the individual and who is the donor for gene or code conjugations. These decisions are then sent to the Ooopsi which follows the instructions by deciding on how exactly to execute the variation types, requesting the needed code parts from the donor and executing the variations. There are three main types of variation (gene conjugation, code conjugation and mutation) and several subtypes (gene deletion, gene insertion, code deletion, code insertion and different types of mutation).
Gene conjugation is an exchange of complete genes between individuals. It is divided into the two subtypes gene deletion and gene insertion19. As the name implies, gene deletion removes genes from the individual genome. On the basis of chance and probabilities that are adjustable by parameters, Dcomm.vary() decides if gene deletion is to be applied and if yes how many and which genes are to be taken away. Analogously, it decides on gene insertion20 with additionally deciding about the donor individual for the inserted genes.
Code conjugation exchanges parts of the evolvable code of two distinct genes. If the donor gene is part of an individual in another Ooopsi, this gene also has to be requested for transfer from this Ooopsi by the host Ooopsi of the varied individual. Like gene conjugation, code conjugation can be divided into two subtypes code deletion and code insertion. The same reasons for this apply as for the two subtypes of gene conjugation. Code conjugation most closely resembles classical GP crossover, because it can choose arbitrary parts of the code for the mixture. In contrast, gene conjugation differs from classic GP crossover in that the boundaries of the exchanged parts (which correspond to the crossover points) are given by the division of the program into separate genes. These boundaries are not a product of chance in the crossover process, but their content evolves21. This is a reason why the division into genes realized in object-oriented ontogenetic programming is a promising technique for finding good building blocks that can be combined by gene conjugation into a better overall program22.
In the first experiments with OOOPS, code conjugation was not yet used. Perhaps, it is not even needed at all23. One could argue with Kevin J. Lang and Peter J. Angeline [Lang, 1995,Angeline, 1997] that the classical GP crossover is not more than a macromutation operator that does not help in finding and reusing good building blocks but only makes big changes to the code and as such could be an important part of GP in providing big jumps through the search space. But in OOOPS, classical crossover (resp. the corresponding code conjugation) or another form of more explicit macromutation is perhaps not needed because of two reasons:
But the OOOP paradigm implies some more variable parameters of the genes that do not exist in classical programming but are central determinants of the behaviour of object-oriented ontogenetic programs. These are the message production of the gene (which consists of the message type and the intensity of production) and its execution conditions. In OOOPS, the execution conditions are divided into requirements and inhibitors (which both also consist of a message type and an intensity). Requirements define a concentration of a specific message that must be reached at the cell's position for executing the genetic code. Inhibitors in contrast to this define the concentration of a message that must not be reached if the gene is to be executed. All these parameters can be mutated by exchanging either the type or the intensity by a random new value (within predefined limits)25.
In OOOPS, all types and possibilities of variation have their own probabilities and can all be applied to the same individual if all happen to be chosen. This also enables a sort of macromutations, but the single probabilities should normally be chosen so that in average not much more than one mutation type at a time is applied.
A typical evolutionary run with OOOPS proceeds as follows:
OOOPS is a big system with many features of which some have been described in the last section. Now we will take a closer look at some features of OOOPS (or OOOP) that are new to genetic programming or very rarely realized until now. Table 3.1 gives an overview of the most important innovations that will be discussed in the following.
|
Genetic programming did get its name from genetics (because of the operators), but until now it did usually not include genes. As we know, genes are the units in the genotype that describe a separate part of the functioning of the cell (a protein). In crossover, genes are normally not split. We also know that genes can be activated or inactivated. Separate parts of the program that have these genetic attributes are introduced by combining genetic programming with OOOP. We already discussed the advantages of using gene regulation in section 2.1.3. But separate genes might also have another advantage for GP that was already mentioned in section 2.1.1: genes are a new approach to modularization of programs.
Genes as separate blocks of code that can evolve through mutation and are recombined into different individuals by gene conjugation are a new technique for evolving reusable program modules. One might argue that only by having the ability to migrate into other individuals and influencing their fitness, which in turn influences the spread of the individual (and with that of the gene) in the population, the gene will evolve towards being a good reusable building block. As genes can spread independently in the population through gene conjugation, it is plausible that not only good individuals but also good genes have a higher probability of survival than bad genes. This implicit genetic evaluation and selection is supported in OOOPS by adding an explicit gene fitness and favouring fitter genes for gene insertion. This has already been described in section 3.2.1 and we will also come back to it later in this section.
In genetic programming, introns27 (or substitutions like explicitly defined introns [Nordin et al., 1996]) have been used as emergent borders for good building blocks. They might also play such a role in nature. But nature additionally has other (more important) mechanisms for defining the borders of a gene. Using introns or their substitutions as emergent borders takes much computing resources during execution of the program or for extracting the intron code before execution or for the genetic operators (in the case of explicitly defined introns).
By using explicit separate genes like in OOOP in which the code evolves, we have predefined borders but emergent content. This has many advantages. First, the crossover operator is very simple, because it only has to mix the genes. It does not have to take care of anything else, neither of producing valid programs nor of computing the probabilities for different crossover points. Second, this simplicity of the crossover operator is not only easy to program but also saves computational resources. Third, this allows to very easily identify and transfer the building blocks between several computers for distributed evolution. Fourth, it enables the use of gene regulation whose pros have been extensively discussed in section 2.1.3. I believe that this technique is also more effective in finding good building blocks than previous techniques (among other things because it enables to apply multilevel evolution as discussed later in this section). This remains to be proved.
Compared to other modularization techniques like automatically defined functions and automatically defined macros, the use of genes in OOOPS does not require to predefine static parameters like for example the number of genes. We have already talked about these advantages of OOOP in section 2.1.3.
As we have seen in section 3.2.1, the object-oriented ontogenetic programming system is what I call an asynchronous GP system. This means that the GP individuals reach the different states of the evaluation-selection-variation loop not at the same time. Their life-cycles are chronologically independent of each other. The term "steady-state" refers to the fact that in an asynchronous GP system the biggest part of the population usually is not also just being varied when one individual changes.
Until now, steady-state approaches are intimately tied to tournament selection, because this is the only known selection method that does not compare the evaluation results of all individuals in the population. With tournament selection, the individuals taking part in the tournament can be selected independently of the rest of the population. Tournament selection is a good selection method. It has many advantages on the other methods: A central unit for selection is not needed, there is no need for a global fitness function that produces absolute fitness values28, the evaluation and selection takes less computational resources, this selection method is closer to the natural example than the others, etc. But tournament selection also has some disadvantages:
As you already know from other sections (e.g. 1.3 and 3.2.1), OOOPS uses conjugation instead of crossover. This decision was made because conjugation only changes one of the two participating individuals which is ideal for asynchronous genetic programming. In the asynchronous approach we always only have one individual which is guaranteed to be at a specific state of the evaluation-selection-variation loop. When this individual is to be varied, with conjugation it can be chosen to get parts of any other registered individual regardless of the state of the last. But if crossover was to be used, the first individual would have to wait for the second to also reach the state of variation so that both would be ready for changes.
This is not the first time that conjugation is proposed for use with evolutionary algorithms. Peter W. H. Smith suggested a form of conjugation in [Smith, 1996] for GAs and GP which consists of copying a part of the donor genome to the identical position in the recipient genome replacing the original recipient code at that position. This conjugation only partly resembles the natural example, because in nature, the transferred genes do usually not replace the original genes but are simply added to the recipient genome.
The OOOPS conjugation follows the natural example in this respect by providing gene insertion without deleting the original code. But it also provides gene deletion for being a good replacement for crossover (we discussed this earlier) and for in average not blowing up the code length. The division of conjugation into insertion and deletion enables a great variability in code length with still (by using the same probabilities) not predetermining any direction of length development.
The reproduction management of OOOPS has already been explained in section 3.2.1. An individual which is selected to produce offspring is registered in the parent table of the database. When an individual is selected to die, a random individual from this database table is used as parent of the replacing child and deleted from the table. If there is no entry in the table, a newly initialized individual is used as parent. This organization of the reproduction process is not only perfectly adapted to asynchronous distributed evolution (with a central unit), it also as a by-product has the advantage of refreshing the diversity of the population and trying new starting points for the exploration of the search space from time to time by inserting new random initial individual into the population. This only happens if there are no parents in the database, but as the experiments until now suggest, this is periodically the case. It is an interesting question for further research why this seems not to happen with a random frequency but in periodically changing phases of high and low offspring production. Could it perhaps be compared with periodic changes in natural population developments?
In section 3.2.1, we also already mentioned that new individuals can always be added to the population by starting new Ooopsis. So the population size can be varied in the course of the evolutionary run if desired. This is a good and important quality for distributing the evolutionary runs on the internet, because new users can always join in and start an Ooopsi on their computer. Yet, distribution of OOOPS on the internet would require some changes in the communication because Ooopsis then regularly change their address and are often unreachable. Consequently, it would make more sense to not let the Ooopsis communicate directly with each other but instead only let them talk to the Ooopse. This means that all individual sourcecodes should also exist in the Ooopsed database35. Moreover, some security features would have to be added.
As mentioned earlier, OOOPS not only determines fitness values for the individuals but also for the genes. Not only the individuals are selected, but also the genes underly a separate selection process which depends on their fitness. Genes are selected for being the parent of inserted genes in gene conjugation depending on their fitness rank. Of course, they are also varied (by mutation and possibly code conjugation). So not only individuals are evolving in the evaluation-selection-variation loop. Also their subunits (genes) follow these steps. This means that we have two levels of evolution in OOOPS. Such multilevel evolution is not unnatural. In fact, we can view many systems on very different levels in nature as evolutionary systems. Elliott Sober writes in [Sober, 1992] about this issue:
...If an object and its offspring resemble each other, the system will evolve, with fitter characteristics increasing in frequency and less-fit traits declining.
This abstract skeleton leaves open what the objects are that participate in a selection process. Darwin thought of them as organisms within a single population. Group selectionists have thought of the objects as groups or species or communities. The objects may also be gametes or strands of DNA, as in the phenomena of meiotic drive and junk DNA.
Outside the biological hierarchy, it is quite possible that cultural objects should change in frequency because they display heritable variation in fitness. If some ideas are more contagious than others, they may spread through the population of thinkers. Evolutionary models of science exploit this idea. Another example is the economic theory of the firm; this describes businesses as prospering or going bankrupt according to their efficiency.
Genes in OOOPS have their own selection and they also reproduce. But they do not reproduce independently of their hosts because their fitness strictly depends on the individual fitnesses36. As has been mentioned in section 2.1.3, this is an important precondition for cooperation between the genes to emerge. They must capture all effects that their behaviour has on others. So the genes in OOOPS evolve separately, but they evolve to cooperate. This type of evolution is called cooperative coevolution.
All environmental influences on the individual are defined in the individual itself, either with the external message sources that feed the externally modelled environment into the individual or with the internal environment model in the distributed problem solving approach of OOOP37. Also the whole functioning of ontogeny with the virtual space and the organization of diffusion and gene regulation is implemented in the individual. These parts of the individual are normally not evolvable because some are not part of the genome and therefore not really constitute the genotype of the individual (which is the only thing usually varied in artificial evolution) and they all do not correspond to attributes of natural genomes that change during the evolution of natural species38. But because they are part of the OOOP individual, they can also be evolved with OOOPS. A very simple example would be that you initialize half of the population with individuals that update its cells in a prespecified and static order and the other half of the population with individuals that randomly choose the next cell that is to be updated. This difference does not exist in nature, but it can make a great difference in the behaviour of the object-oriented ontogenetic program. In the course of the evolutionary run, the individuals whose update method produces better results will supersede the others. This example is a very simple form of evolution, because the search space only contains two points. But it shows that OOOPS also allows to evolve parameters of the multicellular model itself and not only the genome of the modeled cell cluster. Such an evolution of the model can be viewed as a kind of metaevolution.
Nature uses some tricks to reduce the probability that crossover will have disastrous effects on the individual by destroying or removing important genes. Two important such tricks are speciation and homology. Both limit the possibilities of crossover on different levels. Speciation only allows individuals of similar species to produce offspring together. As crossover only happens with sexual reproduction, this means that crossover only happens between the genomes of similar species (which have similar genomes). Homology describes the fact, that crossover tends to exchange very similar parts of the genetic code between the two genomes which mostly serve the same function. This results in quite small changes and greatly reduces the probability to remove code parts providing important functions from a genome. It is very difficult to model this on the computer for enhancing evolutionary algorithms, because the similarity of function between genome parts cannot easily be measured. Nature apparently also does not measure function but in natural crossover, the similarity of the DNA sequence seems to be a sufficient indication of function for providing the advantages of homology. This usually does not hold in evolutionary computation. There has been some work on trying to model homology for GP either by only defining homologous code as being found at the same position in the genome [D'haeseleer, 1994,Altenberg, 1995] or by introducing a much more sophisticated definition of homology by comparing both structure and function of the code as proposed in [Banzhaf et al., 1998]. Though the results did not yet indicate a great breakthrough, it might be a good idea to try some kind of homology on OOOP.
One approach to homology for OOOP could be to introduce a genealogy for genes. Provided a good datastructure for storing the sequence of all past IDs of a gene, one could easily determine the genealogical distance between any two genes. You only would have to find the last ID common to both lines and add the number of IDs following this common ID including the actual gene IDs. In case of no common ID, the genes would not be related. As a gene changes its ID with every variation, this genealogical distance would provide a very easy measure of structural similarity between two genes39.
In the current implementation of OOOPS, homology makes no sense because insertion and deletion are separate variation operators. Homology can only enhance genetic operators in which a part of a genome is replacing a part of another genome40, because only then these two parts can be compared. But the only thing one has to change to enable homology for gene conjugation in OOOPS is to exchange gene insertion and gene deletion with a single gene conjugation operator which replaces a recipient gene by a donor gene. Then you can ensure some kind of homology by only allowing gene conjugation to exchange genes with a genealogical distance below a certain limit.
If you would analogously combine code insertion and code deletion into a single code conjugation, you could even view this genealogically limited code conjugation as a form of speciation for genes. A genealogical distance below the specified limit would mean that the genes are of the same "gene species" and can therefore be recombined.